Tutorial 3: Graphs
Graphs: The art of designing information
Week of September 16, 2024
(3rd week of classes)
How the tutorials work
The DEADLINE for your report is always at the end of the tutorial.
The INSTRUCTIONS for this report is found at the end of the tutorial.
While students may eventually be able to complete their reports without attending the synchronous lab/tutorial sessions, we strongly recommend that you participate. Please note that your TA is not responsible for providing assistance with lab reports outside of the scheduled synchronous sessions.
The REPORT INSTRUCTIONS (what you need to do to get the marks for this report) is found at the end of this tutorial.
Your TAs
Section 0101 (Tuesday): 13:15-16:00 - Aliénor Stahl (a.stahl67@gmail.com)
Section 0103 (Thursday): 13:15-16:00 - Alexandra Engler (alexandra.engler@hotmail.fr)
General Information
Please read all the text thoroughly; do not skip any sections. The tutorials are centered around R exercises designed to help you practice running statistical analyses and interpreting the results. This tutorial will help you develop basic to intermediate plotting skills in R. It is based on code adapted from Chapter 2 of ‘The Analysis of Biological Data,’ with added details and features to enhance your understanding of various R graphic functions.
Note: Once you get used to R, things get much easier. Since this is your 3rd tutorial, try to remember to:
- set the working directory
- create a RStudio file
- enter commands as you work through the tutorial
- save the file (from time to time to avoid loosing it) and
- run the commands in the file to make sure they work.
- if you want to comment the code, make sure to start text with: # e.g., # this code is to calculate…
General setup for the tutorial
An effective approach is to have the tutorial open in our WebBook alongside RStudio.
Bar graphs
1st bar graph
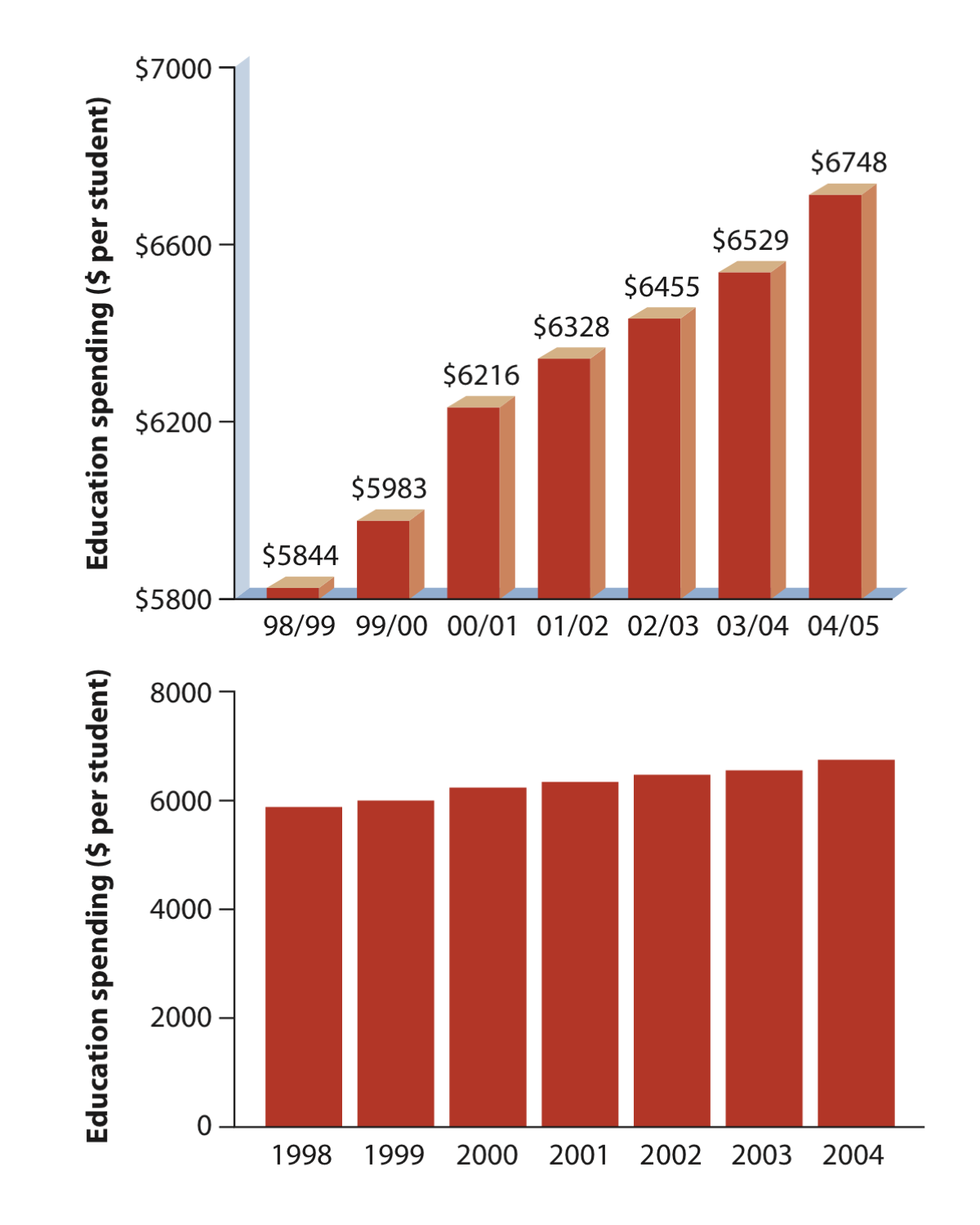
The simplest bar graph possible: This is the education spending data presented in our lecture 3. The data do not need to be summarized and all we will do it to plot the data spending in education through time.
Download the spending data file
Now open the csv file (e.g., in excel) and try to understand how the data are organized.
Now load the data in R as follows:
educationSpending <- read.csv("chap02f1_3EducationSpending.csv")
head(educationSpending)
View(educationSpending)Remember that the function head plots the data for the first 6 observational units. As you noticed, data matrices are usually organized as having different variables as columns. So in the education spending data, variables year and spendindPerStudent are in two different columns. Again, there are several ways to access values in a particular variable (column) in a matrix separately, but the easiest is to use the $ (dollar) sign where the name of the data matrix preceeds the $ sign and the name of the variable of interest succeeds the $ sign. Type and run the code:
educationSpending$year
educationSpending$spendingPerStudentTo produce the barplot, then:
barplot(educationSpending$spendingPerStudent, names.arg = educationSpending$year)You will notice that the barplot produced is not as nice as the one in the Whithlock and Schluter book (see lecture 3) as below:
We can do much better by using and changing arguments within the function barplot as well as using other functions. Let’s start by adding the X-axis and Y-axis labels:
barplot(educationSpending$spendingPerStudent, names.arg = educationSpending$year,
ylab = "Education spending ($ per student)",xlab = "year")This is better but still not great! We will learn how to plot better in the next graph.
2nd bar graph
Influence of serotonin (the ‘happy chemical’) on the social behaviour of desert locusts (as discussed in Lecture 3). The data shows the serotonin levels of each individual, measured separately over time. Here, time serves as the experimental treatment, representing the duration each individual spent in a crowded environment with others.
Download the seratonin data file
First open the file “chap02f1_2locustSerotonin.csv” in excel and try to understand how the data are organized.
Now load the data in R:
locustData <- read.csv("chap02f1_2locustSerotonin.csv")
View(locustData)The bar graph is based on the mean values per treatment (control, 1 hour, and 2 hours) as shown below:

Mean values per treatment can be calculated as follows:
mean.per.treatment<-aggregate(locustData$serotoninLevel,list(locustData$treatmentTime),mean)
mean.per.treatmentThe function aggregate selected the data that belong to the same categorical value in a list (here Treatment, i.e., 0, 1 or 2) and applied a function on these values separately. Here we calculated the mean per treatment, hence mean as the third argument in the function aggregate; we can easily calculate the median by replacing mean by median as follows:
median.per.treatment<-aggregate(locustData$serotoninLevel,list(locustData$treatmentTime),median)
median.per.treatmentLet’s produce the bar graph based on the means:
barplot(mean.per.treatment$x, names.arg = mean.per.treatment$Group.1,xlab = "Time Treatment",ylab = "average seratonin (pmoles)")Keep in mind that label names can be customized as needed, but they should accurately represent the problem at hand. Notice that the current graph is not as clear as the one lecture 3.
To improve this, let’s first set the Y-axis to range between 0 and 12, as shown in the book, so that the bar does not exceed the Y-axis in the current version of the graph.
barplot(mean.per.treatment$x, names.arg = mean.per.treatment$Group.1,xlab = "Time Treatment",ylab = "average seratonin (pmoles)",ylim=c(0,12))Still needs some work though. Let’s change the colour of the bars:
barplot(mean.per.treatment$x, names.arg = mean.per.treatment$Group.1,xlab = "Time Treatment",ylab = "average seratonin (pmoles)", ylim=c(0,12),col="firebrick")There are different colour names for R plots. They can be found at: http://www.stat.columbia.edu/~tzheng/files/Rcolor.pdf
There is one last touch missing; the horizontal line under the bars; now it will look just like the one in the book; h is an argument in the function line that stands for horizontal.
abline(h=0)Of course, we don’t need to go through each of these steps individually; we could have accomplished everything with a single line of code. However, learning step-by-step is much easier while we are still beginners in R:
barplot(mean.per.treatment$x, names.arg = mean.per.treatment$Group.1,xlab = "Time Treatment",ylab = "average seratonin (pmoles)",ylim=c(0,12),col="firebrick")
abline(h=0)3rd bar graph
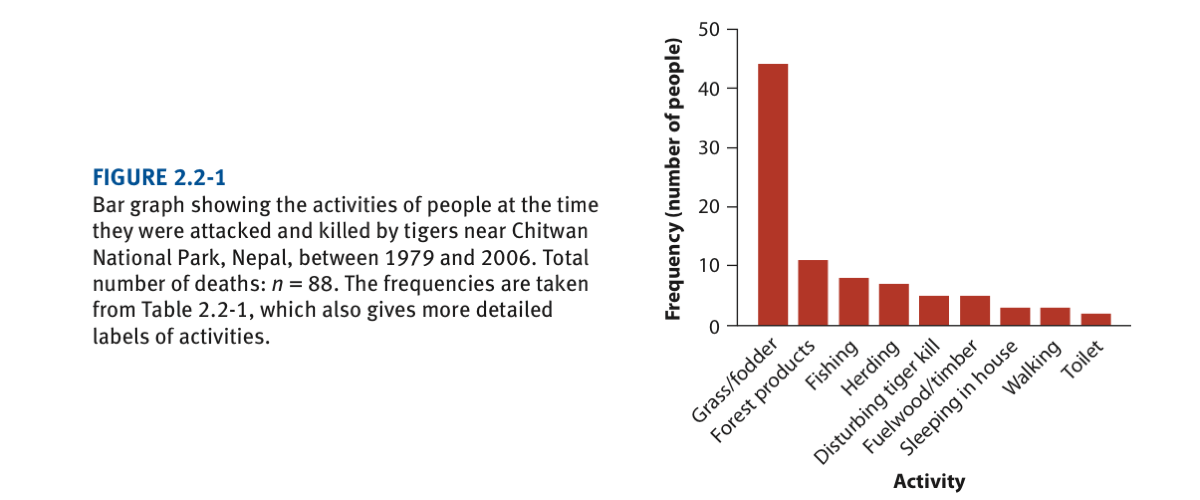
Death by tiger attack acccording to activities at the time they were attacked (see lecture 3). The graph is as follows and we will try to reproduce it very closely:
Download the death by tiger data file
First open the file “chap02e2aDeathsFromTigers.csv” in excel and try to understand how the data are organized.
Now load the data in R:
tigerData <- read.csv("chap02e2aDeathsFromTigers.csv")
View(tigerData)
head(tigerData)Note here that the data is in the form of the activity per person. Given that we want to present the number of people that were killed per activity, we need to calculate these values. The function table counts the number of times that each activity is recorded:
tigerTable <- table(tigerData$activity)
tigerTableWe should then order (sort) so that the bars go from the largest number of individuals to the smallest per activity:
sorted.tigerTable <- sort(tigerTable, decreasing = TRUE)
sorted.tigerTableWe could had performed the two operations above in one single line of code:
sorted.tigerTable <- sort(table(tigerData$activity), decreasing = TRUE)
sorted.tigerTableNow let’s plot the bar graph:
barplot(sorted.tigerTable, ylab = "Frequency", cex.names = 1, las = 2)The graph above has some issues, the most notable being that the X-labels (i.e., activities) are unreadable. To address this, we need to adjust several arguments and introduce new commands related to plot functions.
The first thing we need to know, is to change the plotting area. Function par allows one to enter graphic parameters. mar is an argument of the function par that allows to change the margin sizes of the plot region (i.e., bottom, left, top and rigth margins of the plot region).
par(mar=c(10,6,10,6))
barplot(sorted.tigerTable, ylab = "Frequency", cex.names =1, las = 2,ylim=c(0,50),xlab="activity")When using graphic functions such as par, often other graphs are also affected; to reset R to its graphics default, we use the function dev.off() without any argument inside the parentheses, i.e., just ():
dev.off()Note that the X-label ‘activity’ overlaps with the X-axis, so we need to adjust its position. It can be challenging to remember all these arguments. One helpful approach is to use a Google search to find the information you need. Once you find it, save it in a file to create your own reference guide or ‘cheat sheet.’ For example, you could use the search string ‘label barplot overlap R’ to find the solution. After browsing a couple of links, the answer can be found here:
So, the function mtext (Write Text into the Margins of a Plot) does the trick as follows:
par(mar=c(10,6,10,6))
barplot(sorted.tigerTable, ylab = "Frequency", cex.names =1, las = 2,ylim=c(0,50),xlab="")
mtext(text="activity", side=1, line=8)The final touch (at least for this graph in this tutorial), the line for the X-axis:
abline(h=0)We could experiment further by adjusting label sizes, rotating labels as shown in the book’s graph, changing bar colors, and more. However, it’s time to move on. As we progress, you’ll encounter additional functions and arguments that will help you create more refined and elegant graphs!
Reset graphics to its default parameters so that they won’t interfere:
dev.off()Histograms
Keep in mind that a histogram is a graphical representation of a frequency distribution. By default, R uses right-closed (left-open) intervals for histogram bins. Although this setting can be changed, we won’t cover that adjustment here.
1st Histogram
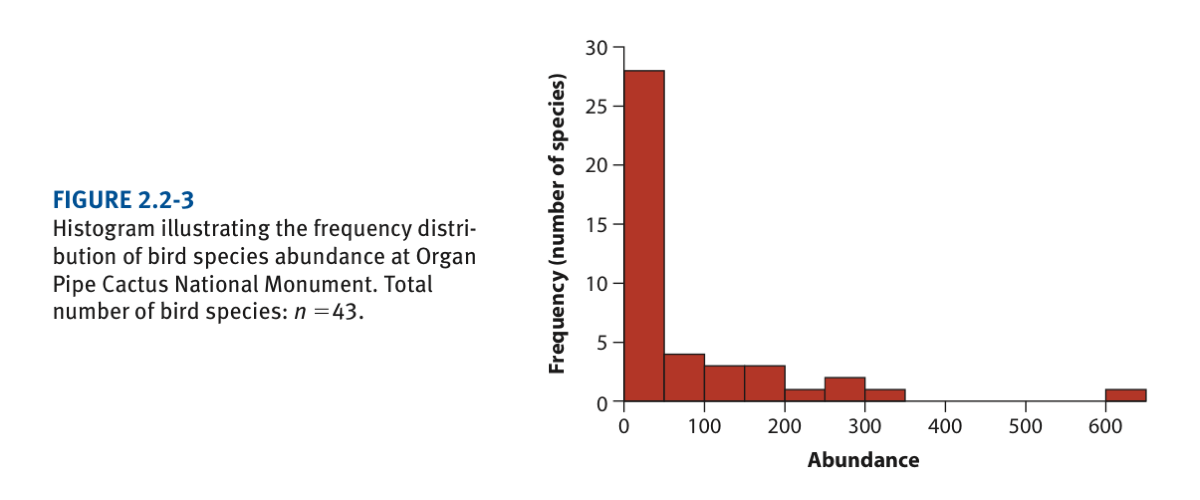
Here we will use the data on the abundance of desert birds (Chapter 2)
Download the bird abundance data file
First open the file “chap02e2bDesertBirdAbundance.csv” in excel and try to understand how the data are organized.
Now load the data in R:
birdAbundanceData <- read.csv("chap02e2bDesertBirdAbundance.csv")
View(birdAbundanceData)
hist(birdAbundanceData$abundance)This looks quite crude compared to the one in the book:
But we will learn how to make it look better in the next example:
2nd Histogram
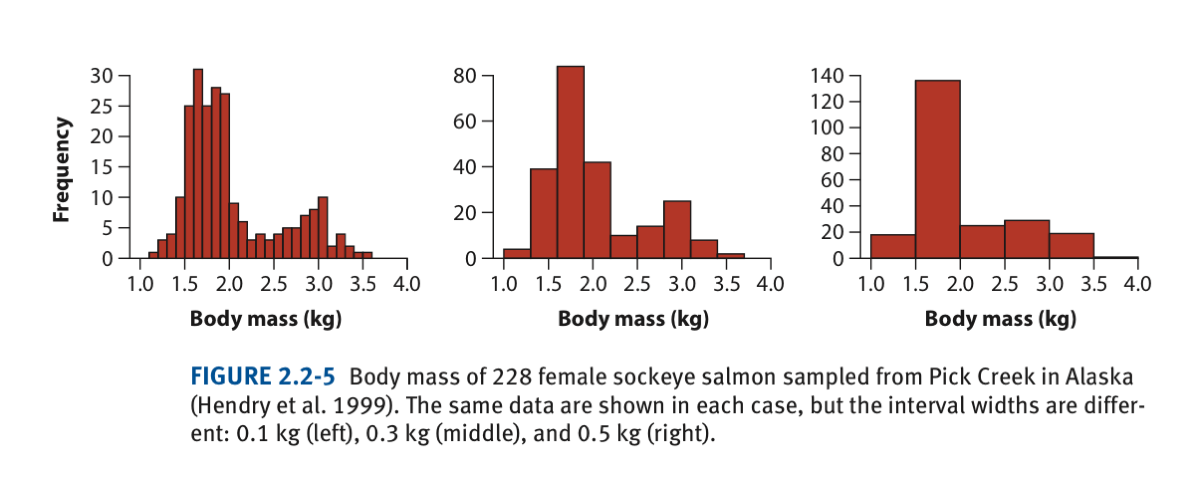
Here we will use the data on body mass (Kg) of 228 female sockey salmon introduced in lecture 4 to produce the following histogram:
First open the file “chap02f2_5SalmonBodySize.csv” in excel and try to understand how the data are organized.
Now load the data in R:
salmonSizeData <- read.csv("chap02f2_5SalmonBodySize.csv")
head(salmonSizeData)In the original histogram found in the lecture and book (shown above), you will notice that the X-axis vary from 1.0 to 4.0 kg; to follow this format, we need to fix the limits of the X-axis and change the by how much (say by 0.1kg or 0.3kg) body mass classes change. This will establish the number of breaks and keep the limits of the X-axis equal between different histograms having different number of body size classes. This is done by using the function seq in the argument breaks in the function hist. Try one histogram at the time
hist(salmonSizeData$massKg, breaks = seq(1,4,by=0.1), col = "firebrick",xlab="Body mass (kg)")
hist(salmonSizeData$massKg, breaks = seq(1,4,by=0.3), col = "firebrick",xlab="Body mass (kg)")
hist(salmonSizeData$massKg, breaks = seq(1,4,by=0.5), col = "firebrick",xlab="Body mass (kg)")Before making one of this histograms look even better, let’s see what the function seq does. When using as an argument for the “break” in the function hist, it establishes the classes, here using 0.1, classes will be:
seq(1,4,by=0.1)In this way it will count how many individuals have a body mass between 1.0 and 1.1., between 1.1 and 1.2, and so on until count the number of individuals between 3.9 and 4.0
Let’s choose one of the histograms and make it look better.
hist(salmonSizeData$massKg, breaks = seq(1,4,by=0.3), col = "firebrick",xlab="Body mass (kg)",main="")
abline(h=0)Scatter plots
Last week (tutorial 2) we saw one way in which the function plot works:
x<-1:7
y<-11:17
plot(x,y)Another way that we adopt in this tutorial is:
plot(y~x)The first thing to notice is that both forms produce exactly the same graph. The second thing to notice is that whereas in the first form, x appears first and y appears second separarated by a comma, in the second form y appears first instead of x and they are linked by a tilda. In R, ~ signifies that y is being graphed as a function of x so that there is some sort of formal link (“dependency”) implied between x and y.
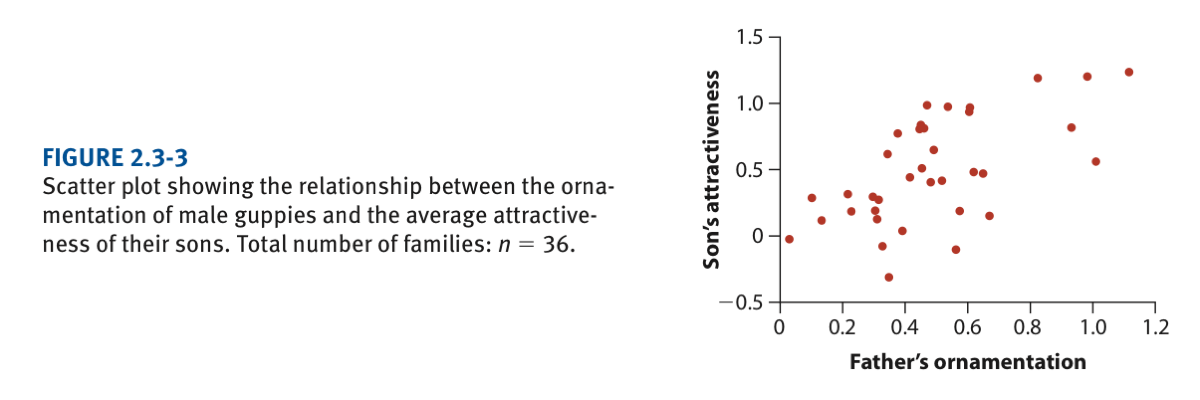
Let’s work on a real example now. Here we will use the data on the relationship between the ornamentation of male guppies and the average attractiveness of their sons (Page 42). The original graph (in the book) is as follows:
Download ornamentation data file
First open the file “chap02e3bGuppyFatherSonAttractiveness.csv” in excel and try to understand how the data are organized. Here, the average attractivness per individual (sons) have already been calculated (n=36) and are the values found in the csv file.
Now load the data in R:
guppyFatherSonData <- read.csv("chap02e3bGuppyFatherSonAttractiveness.csv")
View(guppyFatherSonData)plot(sonAttractiveness ~ fatherOrnamentation, data = guppyFatherSonData)Note then that we are implying here the dependence between the attractiveness of sons (y) and how their fathers are ornamented (x).
The standard plot doesn’t look great. Let’s make it look better using the following steps:
- Make the dots solid:
plot(sonAttractiveness ~ fatherOrnamentation, data = guppyFatherSonData,pch=16)This link shows the codes for all plot symbols (argument pch). Look under “Plotting Symbols” - the number besides each symbol refers to the pch value: http://www.statmethods.net/advgraphs/parameters.html
- Change the colors of the dots:
plot(sonAttractiveness ~ fatherOrnamentation, data = guppyFatherSonData,pch=16,col="firebrick")- Change the labels of the X-axis and Y-axis:
plot(sonAttractiveness ~ fatherOrnamentation, data = guppyFatherSonData,pch=16,col="firebrick",xlab="Father's ornamentation",ylab="Sons's attractiveness")- Change the scales to match the one used in the book (Page 42)
plot(sonAttractiveness ~ fatherOrnamentation, data = guppyFatherSonData,pch=16,col="firebrick",xlab="Father's ornamentation",ylab="Sons's attractiveness",xlim=c(0,1.2),ylim=c(-0.5,1.5))- Finally, remove the box leaving only the 2 axes; As before, one can find this sort of information by searching on the internet. I used the search string “join x and y axis plot R” and found the link https://stackoverflow.com/questions/10874318/r-plot-x-axis-and-y-axis-touching; in there you discover that all you need to do is to set argument bty=‘L’…and voilà!
plot(sonAttractiveness ~ fatherOrnamentation, data = guppyFatherSonData,pch=16,col="firebrick",xlab="Father's ornamentation",ylab="Sons's attractiveness",xlim=c(0,1.2),ylim=c(-0.5,1.5),bty='L')Obviously, the last plot with all the arguments would had produced the desired graph. But, as before, by introducing arguments in steps the different arguments in the function plot helps you understand their characteristics.
Line graphs
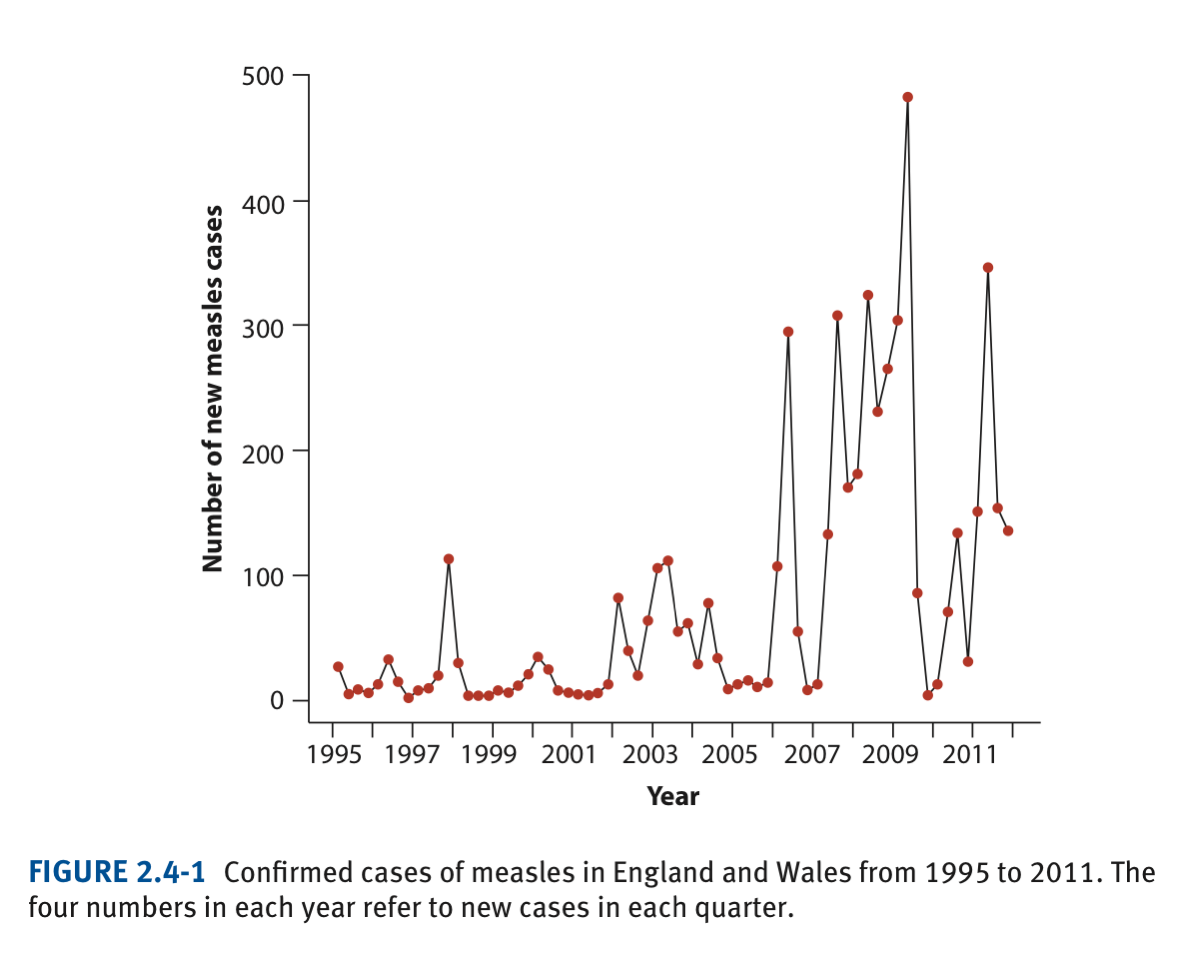
Here we will use the temporal data on number of new measles cases through time (book, Chapter 3 and/or lecture 3). The original graph (from the book) looks like:
Download the Measles data file
First open the file “chap02e4aMeaslesOutbreaks.csv” in excel and try to understand how the data are organized.
Now load the data in R:
measlesData <- read.csv("chap02e4aMeaslesOutbreaks.csv")
View(measlesData)Plot:
plot(confirmedCases ~ yearByQuarter, data = measlesData,xlab="Year",ylab="Number of new measles cases")
lines(confirmedCases ~ yearByQuarter,data = measlesData)Let’s make it look better; change to solid (pch=16) coloured circles (col=“firebrick”). Again, colour codes for R can be at found at: http://www.stat.columbia.edu/~tzheng/files/Rcolor.pdf
plot(confirmedCases ~ yearByQuarter,data = measlesData,xlab="Year",ylab="Number of new measles cases",pch=16,col="firebrick")
lines(confirmedCases ~ yearByQuarter,data = measlesData)Circles are a bit bigger in relation to the book, let’s make them smaller; not it may not look great in the RStudio graph window, but try to Zoom it and you will see how great it looks.
plot(confirmedCases ~ yearByQuarter,data = measlesData,xlab="Year",ylab="Number of new measles cases",pch=16,col="firebrick",cex=0.7)
lines(confirmedCases ~ yearByQuarter,data = measlesData)Grouped bar graphs
Here we have two categorical variables and one numerical. We will use the data reporting reproductive effort and malaria described in chapter 3 (and/or lecture 3). The two categorical variables are the explanatory variable: treatment (control and egg removal) and the response variable: incidence of malaria (yes/no).
Download the bird malaria data file
The numerical values being plotted is the frequency of birds accross the combination of these two categorical variables
birdMalariaData <- read.csv("chap02e3aBirdMalaria.csv")
View(birdMalariaData)We need to first count how many birds in each of the four combinations of the two categorical variables (Malaria/control, Malaria/egg removal, No Malaria/control, No Malaria/egg removal). The function table can be used for that:
birdMalariaTable <- table(birdMalariaData$response, birdMalariaData$treatment)
birdMalariaTableNow, let’s produce the graph:
barplot(as.matrix(birdMalariaTable), beside = TRUE,col=c("firebrick","orange"))Here is the graph with numerous arguments to make it closely resemble the one in the book… phew, it’s a lengthy and detailed piece of code! At this stage, understanding all the arguments and specifics might be challenging, but with time and practice in R and our tutorials, you will get there. The key takeaway here is that R allows you to create highly customized and visually appealing graphs, but it does require an investment of time. The effort is worthwhile, as your code can be saved and revisited even years later. A great way to learn how each argument affects the graph is to modify their values one at a time and observe the changes. With your code in hand, you have the freedom to experiment as much as you like.
The plot might not fit on your RStudio screen, resulting in the error: ‘Error in plot.new() : figure margins too large.’ If this happens, try resizing the plot window in RStudio by adjusting its outer borders with your mouse.
barplot(as.matrix(birdMalariaTable),
beside = TRUE,
col = c("firebrick","orange"),
legend.text = c("Malaria", "No Malaria"),
ylab = "Relative frequency",
space = c(0.2, 1),
args.legend = list (x = "bottomright", bty = "n", fill = c("firebrick", "orange"),
inset = c(0, 0.8),
xpd = TRUE, border=NA),
ylim=c(0,30),
border=NA) # end of entering all arguments for the function barplot
abline (h=0)Mosaic plot for the Malaria data. Note that we transpose the table (i.e., birdMalariaTable) using the function t which stands for transpose.
mosaicplot(t(birdMalariaTable),
col = c("firebrick", "goldenrod1"),
sub = "Treatment",
ylab = "Relative frequency",
cex.axis = 1.1,
main = "")See what happens when you remove t above. You will notice that the variables will change place and it is not so easy to interpret the results.
Report instructions
Create a fictional biological problem (e.g., an experiment you designed) that can be visualized using a mosaicplot. Clearly identify the response and predictor variables, including their units. Then, generate a dataset with fictional values for your problem and save it as a CSV file. You can use Excel to organize the data and then export it in CSV format.
Next, write an R script that reads the data file and generates the plot. Make sure to include all steps within a single R script file. Use # to add comments in the script to describe your fictional problem. For example:
# I set up a study that .....
# use as many lines as necessary,
# always starting with #We have extensive experience with tutorials, and they are structured to ensure that students can complete them by the end of the session. Please submit both the RStudio file and the relevant CSV file through Moodle. A written report (e.g., a Word document) is not required (just the R file with your code as explained above).