Tutorial 5: Sampling variation
Sampling variation, uncertainty, sampling properties of the mean and standard deviation, the standard error of the mean and confidence intervals
Week of September 30, 2024
(5th week of classes)
How the tutorials work
The DEADLINE for your report is always at the end of the tutorial.
The INSTRUCTIONS for this report is found at the end of the tutorial.
While students may eventually be able to complete their reports without attending the synchronous lab/tutorial sessions, we strongly recommend that you participate. Please note that your TA is not responsible for providing assistance with lab reports outside of the scheduled synchronous sessions.
Your TAs
Section 0101 (Tuesday): 13:15-16:00 - Aliénor Stahl (a.stahl67@gmail.com)
Section 0103 (Thursday): 13:15-16:00 - Alexandra Engler (alexandra.engler@hotmail.fr)
General Information
Please read the entire text carefully, without skipping any sections. The tutorials are centered on R exercises designed to help you practice performing and interpreting statistical analyses and their results.
This particular tutorial will guide you in developing basic to intermediate skills in plotting with R. It is based on code adapted from The Analysis of Biological Data (Chapter 2), with additional details and enhancements to several R graphic functions provided here.
Note: At this point we are probably noticing that R is becoming easier to use. Try to remember to:
- set the working directory
- create a RStudio file
- enter commands as you work through the tutorial
- save the file (from time to time to avoid loosing it) and
- run the commands in the file to make sure they work.
- if you want to comment the code, make sure to start text with: # e.g., # this code is to calculate…
General setup for the tutorial
An effective approach is to have the tutorial open in our WebBook alongside RStudio.
Critical notes for this tutorial
While memorizing R commands is not necessary, general concepts learned in the tutorials may be tested in exams. For example, you may be asked to explain the computational methods used in this tutorial (see below for details).
This tutorial is designed to help you understand the nature and characteristics of sampling variation and sampling distributions. As discussed in previous lectures (7, 8, & 9), sampling distributions are essential for understanding and making statistical inferences.
In this tutorial, we will focus on the sampling distribution of the mean and the sampling distribution of the standard deviation, as these are the most important for statistical inference. It’s important to note that since the square of the standard deviation (\(s^2\)) is the variance, the characteristics of the sampling distribution of the standard deviation also apply to the sampling distribution of the variance. However, their shapes differ, which we will explore later in the course.
TIP: We strongly recommend taking notes, preferably on paper, as you work through the tutorials. This practice will help you engage more deeply with the material, promoting better understanding rather than just typing commands. I assure you that taking notes will also be beneficial during exams, as you’ll need to be familiar with the properties and characteristics of estimators (such as the mean and standard deviation, which are statistics based on samples).
Computational versus analytical approaches to build sampling distributions
To grasp sampling distributions and their properties, we will construct them computationally. Traditionally, in classical statistics, these distributions were derived using calculus (i.e., analytically). The key point is that both computational and analytical methods yield nearly identical results for the concepts covered in this tutorial—and in general. However, the computational approach offers the added benefit of helping you build intuition behind statistical methods, as it directly engages with the sampling properties and distributions of estimators (e.g., mean, standard deviation, variance).
Over 100 years ago, when the foundations of statistical inference based on sampling were first developed, computers did not exist, so calculus was the only method available to build sampling distributions essential for statistical inference. While we still rely on analytical methods in many parts of statistics, computational approaches are becoming increasingly common due to the complexity of modern biological problems.
Again, the main reason we are using a computational approach here, rather than an analytical one, is that it offers a more intuitive way to understand the concepts underlying sampling variation and sampling distributions; and how they are used in statistical inference (later in the course). The analytical approach requires a deeper understanding of calculus and probability. While we will explore these concepts through computation, we will still apply analytical solutions that have existed in many cases, more than 100 years. Keep in mind that both approaches provide the same results, as we will demonstrate in later tutorials.
Traditionally, most basic statistical inferential methods are applied using the original methods developed analytically. Analytical approaches tend to be faster because the theoretical framework is already established, and there’s no need to recreate it each time we apply a particular statistical method for inference, unlike the computational approach we are using here.
Keep in mind that the standard error represents the variability around the sample statistic of interest (e.g., mean, standard deviation, etc.). For now, we will focus on the standard error of the mean, but you can calculate the standard error for any statistic of interest, such as the standard error of the standard deviation, variance, median, and more.
Sampling variation based on a normal distribution
Whether we use an analytical or computational approach, we need to assume a form for the population distribution (i.e., the marginal distribution discussed in last week’s lectures) in order to take samples from it—often referred to as ’drawing samples from a population.
Since the normal distribution is well-characterized (i.e., its properties are clearly defined), we can easily generate (or draw) samples from a normally distributed population using most statistical software, including R and even Excel. Most statistical programs have built-in random sampling generators for common probability distributions such as normal, uniform, and log-normal. However, the normal distribution is the most commonly assumed, as we’ve seen in our lectures. Interestingly, as we will explore here and in future lectures, assuming that the values from a statistical population are normally distributed does not mean that if the original population isn’t normal, the statistical methods we use won’t be effective for inference.
Let’s start by understanding how to create a vector of random values drawn from a normal distribution. All the values generated and recorded in the vector are a single sample.
In statistics, we use the term ‘to draw random values from a normal distribution’ (or any other distribution of interest). Random sampling is assumed because each value is independent of the others (recall the principles of random sampling discussed in Lecture 7). As I often say, there is only one way for samples to be independent (random sampling), but countless ways in which they can be dependent. Assuming random sampling simplifies both analytical and computational solutions. However, if the sampling is not random, the statistical methods used for inference may become biased (we will explore this further later in the course).
In the line of code below, the argument n sets the sample size, the argument mean sets the mean of the population of interest and the argument sd sets the standard deviation of the population (remember again that the standard deviation squared \(s^2\) is the variance of a distribution). A normal distribution is defined by its mean and its variance (or standard deviation). The normal distribution used here will be treated as the true population of interest! As such, the function rnorm allows us to sample from a normally distributed population with a given mean and standard deviation set by us. Let’s draw 100 values (i.e., a sample of 100 observations, i.e., n=100) from a normally distributed population with mean \(\mu\) = 20 and a standard deviation (\(\sigma\)) = 1.
Imagine the normal distribution below represents the height distribution of trees in a forest, where the true population mean height (i.e., for all trees) is 20 meters, with a standard deviation of 1 meter. In the example below, we will randomly sample 100 trees from that forest and measure their heights. Although we are sampling a finite number of trees, the normal distribution itself is assumed to have an infinite number of observational units (i.e., an infinite number of trees). Assuming statistical populations of infinite size is common and works well for finite populations. For analytical and computational purposes, statistical populations are generally considered infinite.
sample1.pop1 <- rnorm(n=100,mean=20,sd=1)
sample1.pop1Let’s plot the frequency distribution (histogram) of our sample and calculate the sample statistics we will be working on today:
hist(sample1.pop1)
mean(sample1.pop1)
sd(sample1.pop1)
var(sample1.pop1) # or sd(sample1.pop1)^2Let’s take another sample from the same population:
sample2.pop1 <- rnorm(n=100,mean=20,sd=1)
sample2.pop1
hist(sample2.pop1)
mean(sample2.pop1)
sd(sample2.pop1)
var(sample2.pop1) # or sd(sample2.pop1)^2Notice how the values for the summary statistics (mean, standard deviation, and variance) varied across samples? Even though they were drawn from the same population, both the data and the summary statistics differ between samples. This illustrates the principle of sampling variation, which we discussed in Lectures 7 and 8.
Let’s now generate a sample from another population with \(\mu\) = 20 and \(\sigma\) = 5 (a larger standard deviation than the value of \(\sigma\)=1 set for the previous population above):
sample1.pop2<-rnorm(n=100,mean=20,sd=5)
sample1.pop2Let’s plot its frequency distribution (histogram) and calculate its mean and standard deviation:
hist(sample1.pop2)
mean(sample1.pop2)
sd(sample1.pop2)
var(sample1.pop2) # or sd(sample1.pop2)^2Just to improve your knowledge of R, let’s plot where the mean value falls in the distribution using a vertical line; the argument lwd controls how thick the line is plotted.
abline(v=mean(sample1.pop2),col="firebrick",lwd=3)Let’s compare the two samples by plotting their frequency distributions (histograms) of each sample on the same graph window using the function par. We will use this function to tell R that the plot will have two rows (one for each histogram) and 1 column, i.e., par(mfrow=c(2,1)). To plot 3 histograms on the same graph window we would set the function par as: par(mfrow=c(3,1)). To plot 4 histograms instead on the same graph window, two on the top row and two on the bottom row, we would set the function par as: par(mfrow=c(2,2)).
Here, we won’t make the histograms look nice as we just want to observe their differences. Note that to be able to observe the differences between the two samples, you need to make sure that the scales of axes x and y (set by the arguments xlim and ylim) are the same:
par(mfrow=c(2,1))
hist(sample1.pop1,xlim=c(0,40),ylim=c(0,50),main="Sample from Pop. 1")
abline(v=mean(sample1.pop1),col="firebrick",lwd=3)
hist(sample1.pop2,xlim=c(0,40),ylim=c(0,50),main="Sample from Pop. 2")
abline(v=mean(sample1.pop2),col="firebrick",lwd=3)Note how both samples are more or less located at 20m (i.e., average of 20m) but that the trees in sample 2 vary much more among each other. We will go back to this issue later on today’s tutorial.
Once finished, set the graph parameters back to their default otherwise you will have problems generated new graphs:
dev.off()From sampling variation to sampling distributions: the case of a population with a small standard deviation
Sampling variation refers to the differences in data and summary statistics that occur from sample to sample, even when drawn from the same statistical population. A sampling distribution represents the distribution of a particular statistic (e.g., mean, variance) across a large number of samples. Analytically, this can be approximated using calculus. Computationally, we can generate it as shown below.
A sampling distribution for any particular summary statistic can be constructed by repeatedly drawing a large number of samples, all with the same sample size (n), from the same population (in this case, a normally distributed one with a given mean and standard deviation; however, other distributions could also be used). We keep the sample size constant because sampling distributions vary depending on the sample size. Essentially, we are mimicking the question: ‘What if I had taken a different sample of the same size from the same population?’
It is very easy to draw multiple samples from any given population. We can use the function replicate to do that. Below we generate 3 random samples containing 100 observations (n=100) each from a normally distributed population with a population mean \(\mu\) = 20 and population standard deviation \(\sigma\) = 1:
happy.normal.samples <- replicate(3, rnorm(n=100,mean=20,sd=1))
happy.normal.samplesNote that the name of the matrix above can take any value. So why not happy.normal.samples?!
The dimension of the matrix is 100 observations (in rows) across 3 samples (each sample is in a different column):
dim(happy.normal.samples)Let’s calculate the sample mean for each sample. For that, we will use the function apply. Each sample is in a different column; as such, we want then the MARGIN=2, i.e., calculate for each column. If we use MARGIN=1, then means are calculated per row of the matrix instead of the column. Finally, FUN here will be the mean.
apply(X=happy.normal.samples,MARGIN=2,FUN=mean)Let’s go big now! Let’s take 100000 samples (one hundred thousand samples). This should not take too long. I usually do one million or more but just to demonstrate the principles we are interested here, 100000 is more than fine.
lots.normal.samples.n100 <- replicate(100000, rnorm(n=100,mean=20,sd=1))Calculate the mean for each sample and then plot the distribution of means.
sample.means.n100 <- apply(X=lots.normal.samples.n100,MARGIN=2,FUN=mean)
length(sample.means.n100)See, there are 100000 sample means in the vector sample means.
The frequency distribution contained in this vector of sample means (sample.means.n100) is called “the sample distribution of means”:
hist(sample.means.n100)
mean(sample.means.n100)
abline(v=mean(sample.means.n100),col="firebrick",lwd=3)Note how small the variation is among sample means! Note also how the majority of them are closer to the true population mean of 20 m. This is because the distribution is symmetric. Finally, notice that the mean of these 100000 samples means is equal (for all purposes) to the population from which their samples were taken (i.e., \(\mu\) = 20)
The minimum and maximum values for the 100000 sample means are:
min(sample.means.n100)
max(sample.means.n100)No sample has a mean that should be smaller than 19.2 or greater than 20.8. Wow!
Now, let’s get back to reality where only one sample is usually the case. Let’s say the first sample was the one that you sampled. And the others are just possible samples not sampled (see lecture 7 for details):
sample.means.n100[1]Your value just above is probably a sample mean very very close to the true population. For instance, assume your sample mean was 19.84 m. So the error from the true population value would had been 0.16 m or only 16 cm (i.e., 20 m - 19.84 m) which is a tiny error given tall trees.
Can we trust this sample mean to make inferences? That depends on the sampling distribution of unobserved sample means—i.e., all the other possible samples you could have taken from the population but did not. If the majority of those samples are close to the true population mean, then your single sample is likely to be close as well. This helps introduce the concept of uncertainty. When most samples are near the true population value, you can have a reasonable degree of certainty that your sample mean is also close to the true population value.
In Lectures 7, 8, and 9, we discussed how the variation within a sample (measured by the standard deviation) can help estimate the confidence in how representative the sample is of the population. This concept is what we will explore further in this tutorial.
Note that the population we considered above has a small mean relative to its standard deviation (CV = 1/20 = 0.05 = 5%); remember that we used a sample size of 100 trees.
Changing sample size. Now, let’s observe what happens when the sample size changes. Suppose we sampled 30 trees instead of 100. Should we be as confident in an estimate based on a sample of 30 trees compared to one based on 100 trees? To explore this, let’s estimate the sampling distribution for a sample size of 30 trees. The population parameters remain the same as before (normal distribution with mean = 20 and standard deviation = 1). The only change is the sample size, now reduced to 30 trees instead of 100:
lots.normal.samples.n30 <- replicate(100000, rnorm(n=30,mean=20,sd=1))
sample.means.n30 <- apply(X=lots.normal.samples.n30,MARGIN=2,FUN=mean)Now let’s plot the two sampling distributions:
par(mfrow=c(2,1))
hist(sample.means.n100,xlim=c(18.5,21.5),main="n=100")
abline(v=mean(sample.means.n100),col="firebrick",lwd=3)
hist(sample.means.n30,xlim=c(18.5,21.5),main="n=30")
abline(v=mean(sample.means.n30),col="firebrick",lwd=3)Note how the sample mean values are around the true population value (20 m). Let’s calculate that; i.e., let’s calculate the mean of sample means.
mean(sample.means.n100)
mean(sample.means.n30)Notice that, for all practical purposes, the sample means are identical to the population mean, a property of sampling distributions of the mean discussed in Lecture 8.
However, more importantly, observe that the sampling distribution (i.e., the histogram of the frequency distribution of sample means) shows greater variation among the sample means for samples of 30 trees compared to those of 100 trees. To quantify this, let’s calculate the standard deviation of the sample means. Since the mean of the sample means is the same for both n = 30 and n = 100, we can directly compare these standard deviations.”
sd(sample.means.n100)
sd(sample.means.n30)What do these values tell us? For n = 100, the standard deviation of the sample means is approximately 0.099 m (~10 cm). This indicates that, on average, a sample mean will differ from the true population mean by about 10 cm—not bad considering the average height of the trees is around 20 m, right?
Which standard deviation is smaller? Clearly, the standard deviation of the sample means based on n = 100 trees is smaller. What does this standard deviation represent? It’s the average difference between the sample mean and the population mean. On average, the sample means based on 100 trees are closer to the true population mean (20 m) than those based on 30 trees. Of course, there may be a few samples from the n = 30 group that are closer to the true population mean than some samples from the n = 100 group, but on average, this is not the case, as the smaller standard deviation of the sampling distribution demonstrates.
Under random sampling, on average, we can be more confident that samples based on n = 100 will be closer to the true population mean than those based on n = 30. In other words, by chance alone, there’s a higher probability that a single sample mean from n = 100 will be closer to the true population mean than a sample mean from n = 30 trees.
If you’re still unsure what the standard deviation above is indicating, consider this: What is the probability that a single sample based on n = 100 will be closer to the population mean than a single sample based on n = 30 trees? The standard deviation of a single sample is sensitive to sample variation among samples. To explore this, let’s first calculate the difference between the sample mean values and the population mean:
diff.n100 <- sqrt((sample.means.n100-20.0)^2)
diff.n30 <- sqrt((sample.means.n30-20.0)^2)
dev.off()
boxplot(diff.n100,diff.n30,names=c("n=100","n=30"))Note, again, that there is far more variation (as expected) when samples are based on n=30 than when based on n=100. How many samples are closer to the population value then?
length(which(diff.n100 <= diff.n30))/100000So, there is about a probability of 68% that a single sample based on n=100 trees will closer to the true population value than based on a sample size of n=30.
Conclusion. Well, one has greater certainty (chance) of any given single sample being closer to the true population value for sample sizes that are relatively large (assuming observations, here trees, were randomly sampled from the population).
Since we typically have only a single sample, we can place more trust in a sample based on 100 trees than one based on 30 trees. Although we’ve drawn this conclusion from a normally distributed population, this principle applies to other types of distributions as well (e.g., uniform, lognormal, etc.).
Accuracy & Precision
Let’s put what we have learned above into the perspective of accuracy and precision.
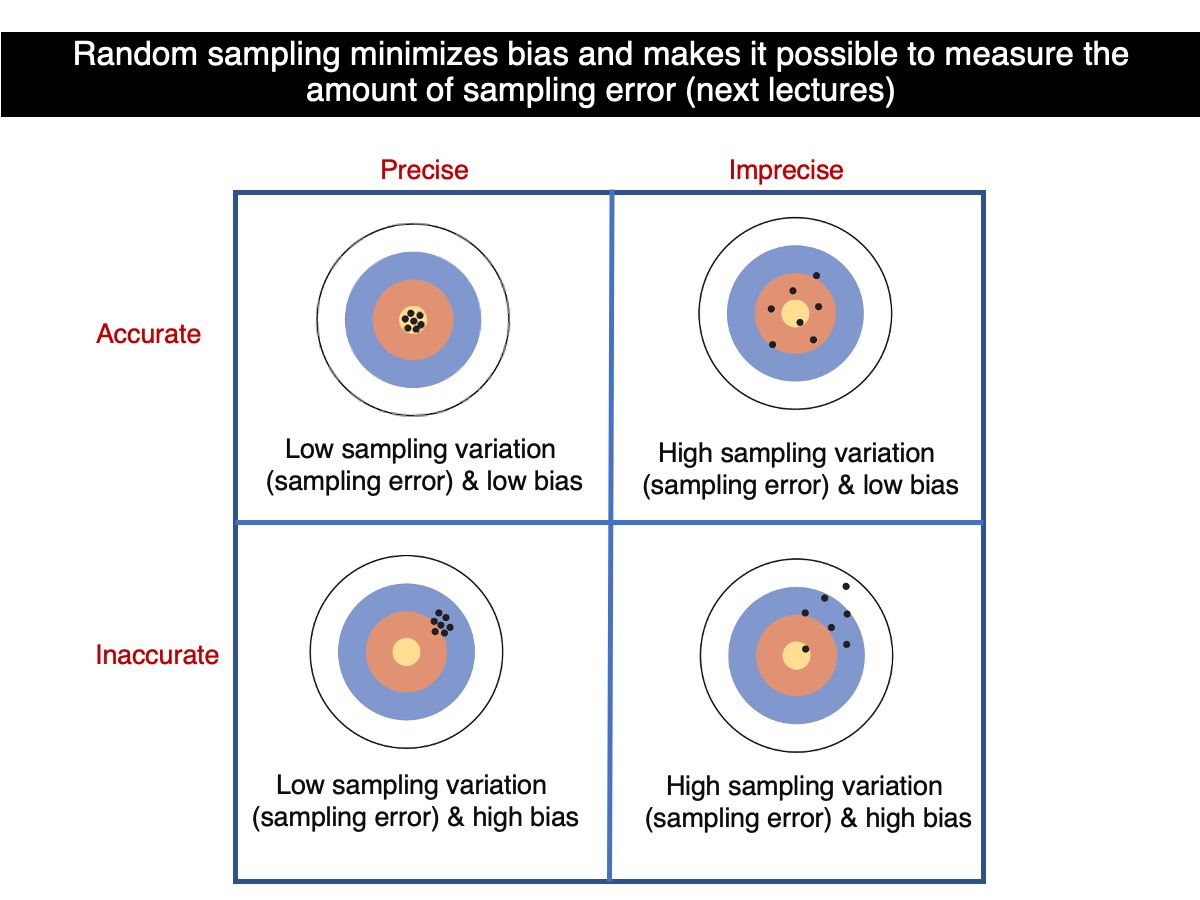
So, which sample size leads to greater accuracy? On average, samples based on 100 trees are more accurate than those based on 30 trees. And which sample size provides greater precision? Samples based on 100 trees, as they tend to be more consistent with one another. Therefore, as sample size increases, both precision and accuracy improve—assuming that random sampling is maintained for all sample sizes.
How to estimate uncertainty from one single sample?
As discussed and demonstrated in our lectures, the variation within samples (measured by the standard deviation of observations) can give us insight into how far sample means may deviate from the true population mean. This is reflected by the standard deviation of the sample means (i.e., the standard error).
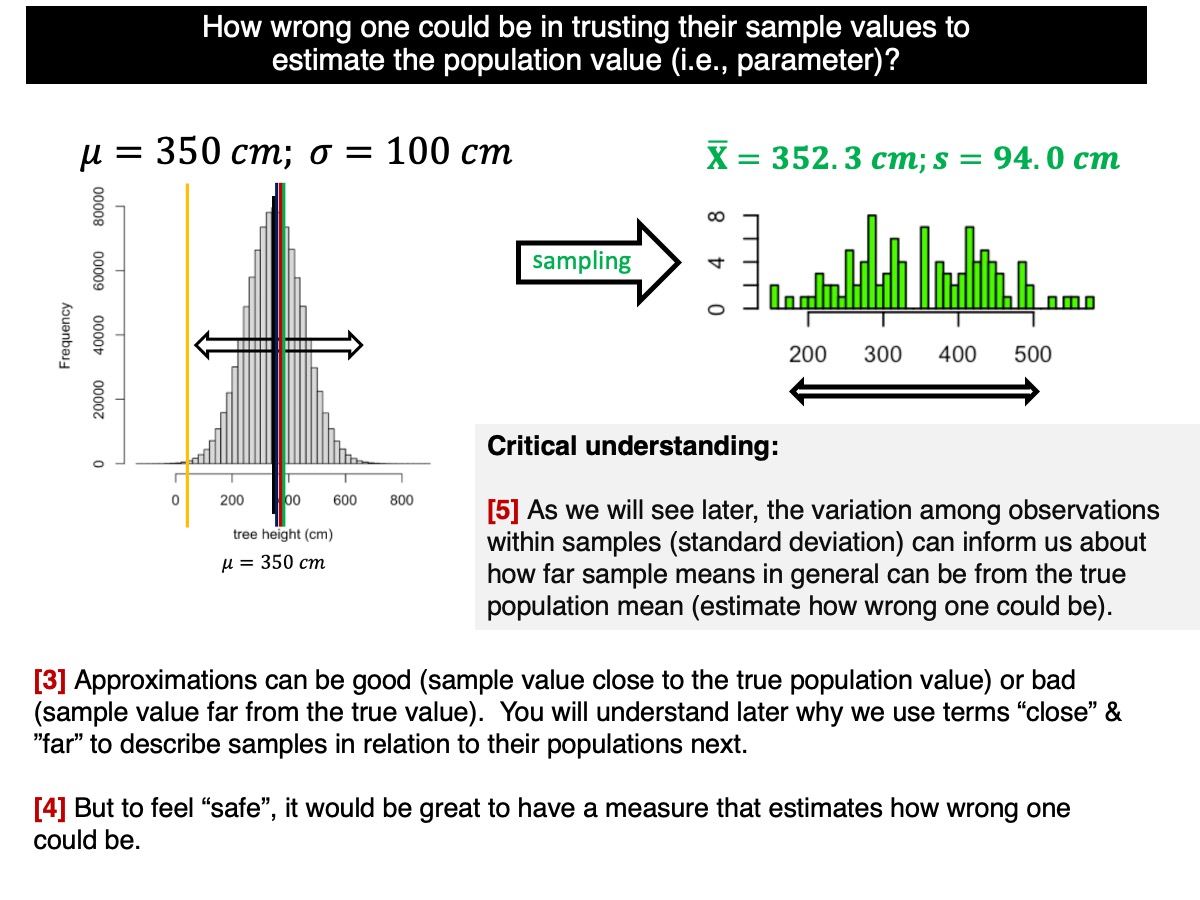
How does that work? Let’s go back to the standard deviation of the sample distributions (i.e., among sample means):
sd(sample.means.n100)
sd(sample.means.n30)Remember, these values are based on the variation of 100,000 sample means around the true population mean. They represent an estimate of the uncertainty (i.e., error) associated with the sample means. The standard error of the sampling distribution of means, denoted as the ‘standard error’ (\(\sigma_\bar{Y}\)), is one of the most important concepts in statistics. It quantifies how accurately (a measure of uncertainty) we can estimate the true population mean. By dividing the population standard deviation by the square root of the sample size, we can calculate the standard error, which should closely match the values obtained above.
The standard error (\(\sigma_\bar{Y}\)) based on a single sample is a statistical estimate of how much sample means would vary around the true population mean if we were to repeatedly sample from the population. A lower standard error suggests that the sample mean is likely close to the population mean, indicating less uncertainty in the estimate. As the sample size increases, the standard error decreases, resulting in smaller confidence intervals and reducing uncertainty in our inference about the true population parameter. Obviously these are statistical (probabilistic expectations). We could have cases in which a small sample could lead to less uncertainty; but this is not the expectation due random sampling (i.e., chance alone). In order words, by increasing sample sizes, we increase the chances of having our sample mean (or any other statistic) closer to the true population mean.
1/sqrt(100) # = 0.1
1/sqrt(30) # = 0.1825They won’t be exactly the same (though pretty much the same) because we used 100000 samples; if we had used 1000000000 the differences would not be perceptible really. If we had then used infinite number of samples (given that the normal distribution as discussed earlier has infinite sizes), then the values would be exactly the same given the correspondent sample size.
All right, so what does that mean? We almost never know the true standard deviation of the population \(\sigma\) either. So how can we estimate how much sampling variation there is around the true population mean \(\mu\)? If the variation is high, then your should be less certain.
What we do though (and it works) is that one can estimate that error based on the standard deviation of a single sample. In other words, we can estimate the standard deviation of the sampling variation around the true population mean \(\mu\) based on the standard deviation of the sample. Wow! That’s statistics for you!
Let’s see how this works! Obviously there is also sampling variation around the true value of the standard deviation. So, let’s estimate standard errors from a single sample and understand how they can estimate how uncertain one can be; remember that lower uncertainty means high certainty (in average).
Let’s go back to the samples based on n=100 trees and n=30 trees. Remember that the mean of sample means equal the true population mean:
mean(sample.means.n100)
mean(sample.means.n30)Let’s now estimate the standard deviation for each sample:
sample.sd.n100 <- apply(X=lots.normal.samples.n100,MARGIN=2,FUN=sd)
sample.sd.n30 <- apply(X=lots.normal.samples.n30,MARGIN=2,FUN=sd)and plot their sampling distributions:
par(mfrow=c(2,1))
hist(sample.sd.n100,xlim=c(0,2),main="n=100")
abline(v=mean(sample.sd.n100),col="firebrick",lwd=3)
hist(sample.sd.n30,xlim=c(0,2),main="n=30")
abline(v=mean(sample.sd.n30),col="firebrick",lwd=3)Boxplots could show their differences even better:
dev.off()
boxplot(sample.sd.n100,sample.sd.n30,names=c("n=100","n=30"))Question - What did you notice? (no need to include the answer in the report)
First, the average of sample standard deviations is equal to the population standard deviation, i.e., 1 here.
mean(sample.sd.n100)
mean(sample.sd.n30)The values above should be between 0.99 and 1.01; i.e., very close to the true population value. The reason (again) that the average of sample standard deviations does not equal exactly the population standard deviation is because we used 100000 samples and not a really huge (or infinite) number of samples. But you get the point!
Second, there is less sampling variation around the true population standard deviation for a large sample size as n=100 trees than n=30 trees.
So, the same patterns of variation related to sample size in terms of precision and accuracy for sample means apply to the sample standard deviation.
Let’s return to our original goal: estimating the uncertainty associated with a single sample. What we are doing here is calculating the error, and based on that, assessing how uncertain we are about whether our single sample provides a reasonable estimate of the true population mean for tree heights. Statisticians use terms like uncertainty and confidence (as we’ll explore throughout the course) rather than certainty. While this may seem like semantics, you’ll come to understand that it reflects a deeper concept—it’s not just a matter of word choice.
Remember, dividing the population standard deviation by the square root of the sample size gives us a measure of the average error (or uncertainty) in how the sample means vary around the true population mean. To estimate this error, we can divide the sample standard deviation of the single sample we have by the square root of the sample size, using this as an estimate of the standard error.
Let’s note that by dividing the sample standard deviations by the square root of their sample sizes, we can estimate the standard error of the sampling distribution for each sample’s mean.
std.error.n100 <- sample.sd.n100/sqrt(100)
std.error.n30 <- sample.sd.n30/sqrt(30)Let’s calculate their means:
mean(std.error.n100)
mean(std.error.n30)No surprises there (well perhaps a bit). The average sample standard error equals the respective population standard errors, i.e., about 0.1 and 0.18.
Let’s put now all this “theory” into practice. Let’s pick the first sample in the series of samples of 100 trees:
sample.means.n100[1]
std.error.n100[1]In my case, which will differ from yours, though the interpretation is the same among every one, the first sample mean based on n=100 trees was equal to 19.92 m and the standard error was equal to 0.103 m (10.3 cm). Remember that the standard error of the mean quantifies how precisely (from a precision point of view) you can estimate the true mean of the population. It tells you that, in average, the sample means will be about 0.103 m (or 10.3 cm) from the true population value. First, 0.103 m is pretty close to the true standard error of the population \(\sigma_\bar{Y}\); and tells us then that, in average, sample means differ from the true population mean \(\mu\) by 0.103 m (i.e., 10.3 cm).
Let’s put this into perspective. It follows then that if we add this estimate of the true average error \(\sigma_\bar{Y}\) (i.e., the sample standard error \(SE_\bar{Y}\) is assumed as a good approximation of the true standard error) around the sample mean value, we should have some expectation that the true population value for the mean \(\mu\) is within that interval. For my sample above, the value would be: 19.92 m - 0.103 m (19.817 m) & 19.92 m + 0.103 m (20.023 m), i.e., 19.92 m \(\pm\) 0.103 m. So, we have some expectation that the true value is in that interval.
The interval above is straightforward to calculate but may be harder to intuitively grasp (though it becomes easier with some understanding of distribution symmetry—something we’ll discuss another time). Let’s explore this computationally: how many of the possible sample means would have an interval that contains the true population mean, \(\mu = 20\) m? Interestingly, the interval we calculated did contain the true population mean. In reality, however, we don’t know whether we’re correct because the true population value is unknown. While we can’t definitively say our interval contains the true population mean, under repeated sampling we have a certain level of confidence that it does (Lecture 9).
Let’s start by building the interval for each sample mean. Remember the one for my sample (first in the vector out of 1000000), was 19.92 m \(\pm\) 0.103 m.
Left.value <- sample.means.n100 - std.error.n100
Right.value <- sample.means.n100 + std.error.n100let’s plot the first 50 intervals:
n.intervals <- 50
plot(c(20, 20), c(1, n.intervals), col="black", typ="l",
ylab="Samples",xlab="Interval",xlim=c(19.5,20.5))
segments(Left.value[1:n.intervals], 1:n.intervals, Right.value[1:n.intervals], 1:n.intervals)A good number of intervals contain (cross) the true population value, i.e., the horizontal bar crosses the vertical bar representing the true population mean value \(\mu\) = 20 cm. In other words that interval contains the true population mean value \(\mu\). But some intervals don’t contain \(\mu\). They are either to the left or to the right. How many in total out of 1000000 intervals contain the true population value? This can be calculated as:
length(which(cbind(Left.value <= 20.0 & Right.value >= 20.0)==TRUE))In my case, there were 68254 out of 100000 that contained the true value of the population. This translates into a chance (probability) of 68.25% that the true population value is contained by one of the sample intervals simply built from sample values, i.e., sample mean \(\bar{Y}\) and sample standard deviation \(SE_\bar{Y}\).
CRITICAL: It’s important to note that this does not mean there is a 68.25% probability that the calculated interval from a single sample contains the true population mean. What it actually means is that 68.25% of the intervals calculated from repeated sampling will contain the true population mean (i.e., the parameter). Any given confidence interval either does or does not contain the true population mean. The probability refers to the overall set of intervals, not a specific one. This is why we refer to it as a ‘confidence interval’—we can be confident that 68.25% of such intervals, based on repeated sampling, will contain the true population mean. Some intervals will be wider and others narrower, but that doesn’t make any one of them more likely to contain the true population mean. As we discussed in our lectures, confidence intervals are crucial but often misunderstood by practitioners.
Note that we did not use any population value in our calculations, since in real life we don’t know the true value. For demonstration purposes here, we simply checked how many of the sample-based intervals contained the true population mean, \(\mu = 20\). Now, let’s consider an interval that’s twice as wide. For my first sample, this would be 19.92 m \(\pm\) 2 x 0.103 m, or 19.92 m \(\pm\) 0.206 m. We are now assuming a margin of error of 0.206 around the original sample mean, which is still quite small (20.6 m) compared to the sample mean of 19.92 m. Let’s now estimate the probability that such intervals contain the true population value:
Left.value <- sample.means.n100 - 2 * std.error.n100
Right.value <- sample.means.n100 + 2 * std.error.n100
length(which(cbind(Left.value <= 20.0 & Right.value >= 20.0)==TRUE))/100000The probability is now much higher, with about 95% of such sample-based intervals containing the true population mean—all estimated using sample data. This is quite good. If, depending on the context, we’re comfortable with an error margin of 0.206 m, we can be highly confident (95% confident) that our interval, 19.92 m \(\pm\) 0.206 m (i.e., 19.714 m to 20.126 m), contains the true population mean. This suggests that our sample mean is a strong estimate of the population mean. However, as mentioned earlier, we still cannot say that any single interval has a 95% probability of containing the true population mean.
In our upcoming lectures and the next tutorial, you’ll learn that the value we multiply by the standard error to estimate confidence intervals is not exactly 2. This value varies with sample size, and we’ll explore this in more detail later. However, using 2 as a ‘rule of thumb’ gives you a good sense of how confidence intervals work and how to estimate uncertainty with a reasonable level of confidence.
This is a long tutorial, covering essential concepts that are critical for truly understanding what we do in statistics. Our course is designed to help you grasp these concepts as we progress.
Report instructions
The report is based on the following problem:
Write code to compare the sampling distribution of means for two normally distributed populations, both with a mean of 25 m. One population should have a standard deviation of 2 m, and the other a standard deviation of 6 m.
Next, answer the question: Which population is more likely to produce a sample mean closest to the true population mean (i.e., the parameter mean)? Use the code you developed to demonstrate your answer.
Focus solely on the sampling distributions—there’s no need to consider confidence intervals. Finally, include a brief explanation of your answer, based on the code and the concepts covered in this tutorial.
# remember text (not code) always starting with #
We have extensive experience with tutorials, and they are structured to ensure that students can complete them by the end of the session. Please submit both the RStudio file through Moodle. A written report (e.g., a Word document) is not required (just the R file with your code as explained above).